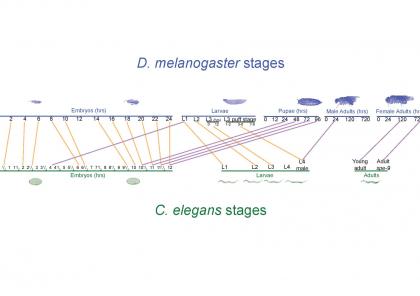
We have developed two new statistical metrics “TROM” (TRanscriptome Overlap Measure) and “EPOM” (EPigenome Overlap Measure) to evaluate the similarity of transcriptomes andepigenomes (i.e., sets of chemical compounds that are not part of the DNA sequence but are on or attached to DNA) within and across species. In work with the modENCODE consortium [1, 2, and 3], we used TROM to discover a previously unknown conservation between the developmental stages of D. melanogaster and C. elegans, two vastly different model organisms that diverged 600 million years ago. We also used TROM to study the conservation of stem cell differentiation across multiple mammalian species [4] and investigated its statistical properties [5]. In [6] we developed EPOM and applied it to large-scale epigenomic data generated by the NIH Roadmap Epigenomics Mapping Consortium that include 12 epigenomic marks and 127 human tissues and cells. EPOM successfully captured epigenomic characteristics of various tissues and cells and discovered previously unknown similarity among them. We demonstrated that TROM and EPOM are more powerful measures than the widely used Pearson and Spearman correlations for establishing sparse correspondence maps of biological samples in terms of transcriptome and epigenome similarities. Moreover, in our ongoing work [7] we show that extending TROM to measure the similarity of genome-wide alternative splicing patterns also reveals meaningful sample correspondence with interesting biological implication.
We also have extensive collaboration with biologists in studies of the binding specificity oftranscription factors (i.e., proteins that bind to specific DNA sequences, thereby controlling the rate of transcription of genetic information from DNA to RNA). Our fruitful collaboration has resulted in work published on high-impact journals including Genome Biology [8] and PNAS [9]. Specifically, our Genome Biology paper is a highly accessed article on BioMed Central and was selected for “Faculty of 1000 Biology”.
References:
[1] Li, J.J., Huang, H., Bickel, P.B., and Brenner, S.E., “Comparison of D. melanogaster and C. elegans developmental stages, tissues, and cells by modENCODE RNA-seq data,” Genome Research 24(7):1086-1101 (2014)
[2] Gerstein, M.B.*, Rozowsky, J.*, Yan, K.K.*, Wang, D.*, Cheng, C.*, Brown, J.B.*, Davis, C.A.*, Hillier, L*, Sisu, C.*, Li, J.J.*, Pei, B.*, Harmanci, A.O.*, Duff, M.O.*, Djebali, S.*, and 82 other authors from the modENCODE consortium, “Comparative analysis of the transcriptome across distant species,” Nature 512(7515):445-448 (2014)
[3] Boyle, A., Araya, C., Brdlik, C., Cayting, P., Cheng, C., Cheng, Y., Gardner, K., Hillier, L., Janette, J., Jiang, L., Kasper, D., Kawli, T., Kheradpour, P., Kundaje, A., Li, J.J., and 25 other authors from the modENCODE and ENCODE consortia, “Comparative analysis of regulatory information and circuits across distant species,” Nature 512(7515):453-456 (2014)
[4] Yang, Y., Yang, Y.C.T., Yuan J., Lu, Z.J., and Li, J.J., “Large-scale mapping of mammalian transcriptomes identifies conserved genes associated with different cell states,” Nucleic Acids Research (2016)
[5] Li, W.V., Chen, Y., and Li, J.J., “TROM: A testing-based method for finding transcriptomic similarity of biological samples,” Statistics in Biosciences (2016)
[6] Li, W.V., Razaee, Z.S., and Li, J.J., “Epigenome overlap measure (EPOM) for comparing tissue/cell types based on chromatin states,” BMC Genomics 17(Supp 1):10 (2016)
[7] Gao, R. and Li, J.J., “Correspondence of D. melanogaster and C. elegans developmental stages revealed by alternative splicing characteristics of conserved exons,” BMC Genomics 18:234 (2017)
[8] MacArthur, S.*, Li, X.Y.*, Li, J.J., Brown, J.B., Chu, H.C., Zeng, L., Grondona, B.P., Hechmer, A., Simirenko, L., Keranen, S.V., Knowles, D.W., Stapleton, M., Bickel, P., Biggin, M.D., and Eisen, M.B., “Developmental roles of 21 Drosophila transcription factors are determined by quantitative differences in binding to an overlapping set of thousands of genomic regions,” Genome Biology 10:R80 (2009)
[9] The ENCODE Project Consortium, “An integrated encyclopedia of DNA RESEARCH elements in the human genome,” Nature 489(7414):57-74 (2012)